ORIGINAL
Genetic structure of Antioquia Holstein from two SNPs and association with dairy traits
Estructura genética del Holstein Antioqueño a partir de dos SNP y asociación con características lecheras
Stephania Madrid G,1* M.Sc, Albeiro López H,1 Ph.D, Julián Echeverri Z,1 Ph.D.
1Universidad Nacional de Colombia, Facultad de Ciencias Agrarias, Departamento de Producción Animal, Grupo de Investigación BIOGEM, Calle 59 A No. 63 – 20, Medellín, Colombia.
*Correspondence: smadridg@unal.edu.co
Received: November 2014; Acepted: March 2015.
ABSTRACT
Objective. Analyze the structure and genetic differentiation of a population of Antioquia Holstein cows from the polymorphisms A192G of INHA and A-320T of FSHR, and explore the association of the genotypic combinations with milk traits. Materials and methods. 1240 lactations of 356 animals from 9 herds in 6 municipalities of Antioquia were analyzed. Genotyping was performed by PCR-RFLP. Structure and genetic diversity parameters were determined using GenAlex software. The association of genotypes combinations with productive and reproductive traits was explored through a linear mixed model. Results. SNP A192G showed a frequency of 0.534 and 0.466 for A and G alleles respectively and SNP A-320T had a frequency of 0.660 far A allele and 0.339 for T allele, this way the population is in HWE. The FST, FIS and FIT values were 0.059, 0.285 and 0.328 respectively indicating a moderate genetic differentiation between subpopulations. The A-320T SNP showed significant effect on milk yield. Fat and protein percentage, calving interval and services per conception were not affected by these polymorphisms or their interaction. Conclusions. Phenotypic selection made on this population has not been strong enough to generate noticeable changes in allele frequencies of these polymorphisms or deviations from Hardy-Weinberg equilibrium. The interaction of these polymorphisms has no significant effect on the characteristics of zootechnical interest, so its use in programs of molecular marker assisted selection is not recommended.
Key words: Dairy cattle, polymorphism, population genetic, SNP-RFLP (Source: AGROVOC).
RESUMEN
Objetivo. Analizar la estructura y diferenciación genética de una población de vacas Holstein de Antioquia a partir de los polimorFISmos A192G de INHA y A-320T de FSHR, y explorar la asociación de las combinaciones genotípicas con características de importancia económica en lecherías especializadas. Materiales y métodos. Se analizaron 1240 lactancias, de 356 animales de 9 hatos en 6 municipios de Antioquia. La genotipificación se realizó mediante PCR-RFLP. Se determinaron parámetros de estructura y diversidad genética utilizando el software GenAlex. La asociación de las combinaciones de genotipos con características productivas y reproductivas se exploró mediante un modelo linear mixto. Resultados. El SNP A192G presentó una frecuencia de 0.534 y 0.466 para el alelo A y G respectivamente y el SNP A-320T tuvo una frecuencia de 0.660 para el alelo A y 0.339 para el alelo T, encontrándose la población en HWE. Los valores FST, FIS y FIT fueron 0.059, 0.285 y 0.328 respectivamente indicando una moderada diferenciación genética entre subpoblaciones. El SNP A-320T presentó efecto significativo sobre la producción de leche. El porcentaje de grasa, proteína, intervalo entre partos y servicios por concepción no se vieron afectados por los polimorFISmos o su interacción. Conclusiones. La selección fenotípica realizada sobre esta población no ha sido lo suficientemente fuerte para generar cambios notorios en las frecuencias alélicas de estos polimorFISmos ni desviaciones del equilibrio de Hardy-Weinberg. La interacción de estos polimorFISmos no presenta un efecto significativo sobre las características de interés zootécnico por lo cual no se recomienda su uso en programas de selección asistida por marcadores moleculares.
Palabras clave: Ganado lechero, genética poblacional, polimorFISmo, SNP-RFLP (Fuente: AGROVOC).
INTRODUCTION
The genetic variability of a population is influenced by the number of founding individuals, genetic drift, strategy and selection intensity (1), while the population structure can be affected by the interchange of genetic material among populations, the reduction of population size, and the decrease in the number of male reproducers, among other factors (2).
The breeding programs implement in developing countries based on importation of genetic material, but the majority of this comes from populations with low genetic variability and high levels of endogamy due to using the few genetically superior individuals that are used for mating, putting at risk the sustainability of the improvement programs (2).
In the dairy herds in Antioquia, artificial insemination is implemented as the principal tool for genetic improvement. Techniques such as multiple ovulation and embryo transfer and producing embryos in vitro are implemented with lesser proportion in order to obtain individuals with high genetic merit that improve the productive levels of those systems.
Among the existing methodologies to select superior individuals we find marker assisted selection (MAS), which are sites in the genome where differences exist in the nucleotide sequence between individuals of the same species, generating single nucleotides polymorphism (SNP) that can be identified by the PCR-RFLP (3). These polymorphisms can also be used to know the structure and diversity of a population (4). The SNP A192G is found in the exon 1 of the inhibin alpha gene (INHA), and since it is a synonymous mutation it does not change the protein amino acid sequence, but rather generates an additional restriction site for the MspI endonuclease (5,6). The SNPA-320T is found upstream from the follicle stimulating hormone receptor gene (FSHR) and although it is not found in any codifying region, it can modulate gene expression and also change the restriction pattern obtained in the Taq1 enzyme (7,8).
The stimulating follicle hormone (FSH) has an essential role in oogenesis, allowing recruitment and follicular development, and acts exclusively through its receptor (FSHR) which is found on the surface of granulosa cells (7,9). Inhibin regulates FSH levels affecting folliculogenesis (5). These polymorphisms have been associated with the response to superovulation and seminal quality, and it has been found that the INHA G allele exercises a positive effect on both traits, while the FSHR A allele improves seminal traits (5,7,9). However, there are no studies on the effect of the principal parameters evaluated in the specialized dairy systems. The objective of this research was to analyze the structure and genetic diversity of a population of Holstein cattle based on A192G and A-320T polymorphisms of INHA and FSHR genes, and to determine the association of its interaction with the main productive and reproductive parameters estimated in the specialized dairy systems.
MATERIALS AND METHODS
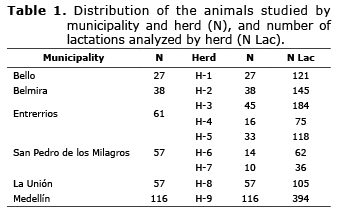
Population study. The phenotypic information and the DNA samples were obtained from 356 Holstein cows (1240 lactation periods) from nine herds in seven municipalities of Antioquia (Table 1).
Analysis of structure and population differentiation. The genomic DNA was obtained from blood samples using the modified Salting out method (10). To genotype the SNP A192G (GenBank number: rs41257116), a fragment of 249 bp of exon 1 was amplified using the primer F: 5’-GCCCTGTTTCTGGATGCC-3’ and R: 5’-ATTCAACCCAACCTGCCTA-3’ (5). The final reaction volume was 30 µl, and 3.8 µl was used as buffer reaction 10X (200 nM (NH4)2SO4, 750 mM Tris-HCl pH 8.8 a 25°C), 2.5 mM of MgCl2, 15 pmol of primer, 0.4 mM of dNTP (triphosphate deoxyribonucleotide), 1.5 units of Taq-DNA polymerase and 100 ng of genomic DNA as a template. The PCR conditions were 5 min at 95°C, 34 cycles of 94°C for 45 s, 62°C for 45s for annealing, 72°C for 45s and 72°C for 10 min for the final extension. Digestion was done with 15 µl of PCR product, 7 U of MspI enzyme and 2.0 µl of buffer in a final volume of 30 µl for 8 hours at 37°C. The amplified fragment contained two common restriction sites and one mutant (GGGAC) for the enzyme in positions 31, 153 and 75, respectively, generating three fragments (123, 95 and 31 bp) for allele A and four fragments (95, 79, 44 and 31 bp) for allele G. The digestion fragments were visualized using electrophoresis in agarose gel at 4%.
To genotype the SNP A-320T (GenBank number: rs43676359) a fragment of 970 bp was amplified with the primers F: 5’-AGTTCGACCGCATCCCTG-3’ and R: 5’-AATTCATTTGTGCCAGCATC-3’ (7). The final reaction volume was 25 µl, and 3.1 µl was used as buffer reaction 10X, 2.7 nM of MgCl2, 5 pmol of primers, 0.24 mM of dNTP, 1.5 units of Taq-DNA polymerase and 100 ng of genomic DNA as a template. The conditions of the PCR were 5 min at 95°C, 35 cycles of 94°C for 30 s, 58°C for 30 s for annealing, 72°C for 30 s and 72°C for 8 min for the final extension. Digestion was done with 12 µl of PCR product, 7 U of TaqI and 1.6 µl of buffer in a final volume 25 µl for 16 hours at 65°C. The amplified fragment contained four restriction sites (T’CG_A) for the enzyme, resulting in five fragments (446, 293, 140, 86 and 5 bp) for A allele. Allele T causes the loss of one of the restriction sites, generating four fragments (586, 293, 86 y 5 bp). The digestion fragments were visualized using electrophoresis in agarose gel at 3%.
The allelic frequencies were determined using formulas ![]() y
y ![]() where NA and Na are the number of A and a alleles, in the population and N is the number of individuals. The genotype frequencies were determined with formulas
where NA and Na are the number of A and a alleles, in the population and N is the number of individuals. The genotype frequencies were determined with formulas ![]() ,
,![]() y
y ![]() where NAA, NAa and Naa are the number of copies of genotypes AA, Aa and aa (11).
where NAA, NAa and Naa are the number of copies of genotypes AA, Aa and aa (11).
Genetic differentiation was determined comparing the observed heterozygosity (Ho) and the expected heterozygosity (He), calculated with GenAlex software (12). The Hardy-Weinberg state of equilibrium (HWE) was determined based on allelic and genotypic frequencies observed and expected using GenAlex software (12).
The population structure was determined using Wright’s F statistics: ![]() ,
, ![]() ,
, ![]() where Ho and He are the observed and expected heterozygosity among subpopulations and HT is the total expected heterozygosity. These were obtained using GenAlex software with the Molecular Variance Analysis (AMOVA) method. The genetic flow expressed as the number of migrants (Nm) was determined with the formula
where Ho and He are the observed and expected heterozygosity among subpopulations and HT is the total expected heterozygosity. These were obtained using GenAlex software with the Molecular Variance Analysis (AMOVA) method. The genetic flow expressed as the number of migrants (Nm) was determined with the formula ![]() (13).
(13).
With the FST values paired among subpopulations, the UPGMA tree was built using MEGA 6 software (14).
The linkage disequilibrium was evaluated using a linkage disequilibrium coefficient which was calculated with GenAlex software using the formula ![]() (11) where fAB is the frequency of the combination of AB alleles and fA and fB is the allelic frequency of A and B respectively; and using the correlation coefficient through sets of loci with the formula
(11) where fAB is the frequency of the combination of AB alleles and fA and fB is the allelic frequency of A and B respectively; and using the correlation coefficient through sets of loci with the formula ![]() (11).
(11).
Association of interaction of INHA and FSHR genotypes with productive and reproductive efficiency. The analyzed parameters were milk yield adjusted to 305 days (PAJUS), protein percentage (PP) and fat (PG) as quality indicators and somatic cell count (SCC) as a health indicator. The analyzed reproductive parameters were calving interval (IEP) and services per conception (SPC). The SCC was transformed to its logarithmic form with the formula SCS=3+log2(SCC/100) (15). To achieve a better balance in data, the births occurring between 1998 and 2003 were grouped in the year 2003; those in 2004 and 2005 were grouped in 2005; and those that occurred between 2011 and 2014 were grouped in 2014. The sixth to the eleventh lactation were grouped in the sixth lactation. The associations of genotypes with the productive and reproductive efficiency of Holstein cows were determined with a Mixed Linear Model. Tukey test was used to determine significant differences between the measurements obtained for fixed effects included in the model. This statistical analysis was done with SAS software (16). The statistical model used was:
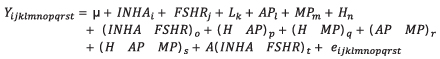
Where Yijklmnopqrst is the dependent variable, which could be PAJUS, PP, PG, SCS, IEP or SPC, µ is the mean for the trait in the population; INHAi is the fixed effect of the INHA genotype [1-3]; FSHRj is the fixed effect of the FSHR genotype [1-3]; Lk is the fixed effect of the lactating number [1-6]; APl is the fixed effect of the birth year[1-8]; MPm is the fixed effect of the birth month [1-12]; Hn is the fixed effect of the herd [1-9]; (INHA*FSHR)o is the fixed effect of the interaction between both genotypes; (H*AP)p is the fixed effect of the interaction between herd and birth year; (H*MP)q is the fixed effect of the interaction between herd and birth month; (AP*MP)r is the fixed effect of the interaction between birth month and year; (H*AP*MP)s is the fixed effect of the interaction between herd, birth year and month; A(INHA*FSHR)t is the random effect of the interaction of the nested genotypes with the animal and eijklmnopqrst is the random error. In addition to these effects, the real production of milk (PREAL) and the length of lactation (DL) were included as covariates in the PP and FT models. For SCS, IEP and SPC, the PAJUS was also included as a covariate, and additionally for SPC the IEP was included, that corresponded to the analyzed lactation as a covariate.
RESULTS
Allelic frequencies and genotypes. The PCR product corresponding to a fragment of the exon 1 of INHA gene presented two common restriction sites and a mutating one for the MspI enzyme generating three or four fragments for the A and G allele, respectively (Figure 1).
The PCR product corresponded to a fragment of the 5’UTR region of the FSHR gene that contained four restriction sites for the TaqI enzyme when the A allele was present, and three restriction sites when the T allele was present, generating five or four fragments, respectively (Figure 2).
For INHA the allelic frequencies were 0.534 and 0.466 for A and G respectively. The genotype frequencies were 0.269, 0.529 and 0.201 for AA, AG and GG respectively. For FSHR, the frequency was 0.660 and 0.339 for the T and A allele respectively. The genotype frequencies found for AA, AT and TT were 0.096, 0.485 and 0.417 respectively (Table 2).
Hardy-Weinberg equilibrium. The total population did not present significant differences among observed (Ho=0.504±0.028) and expected (He=0.466±0.013) heterozygosity for both genes (Table 3). The majority of the subpopulations were found in HWE for both genes, except the subpopulations in Entrerrios and La Unión. The subpopulation in Entrerrios showed a significant reduction in the number of heterozygotes expected for the INHA polymorphism, while the subpopulation of La Unión had an excess number of expected heterozygotes for FSHR polymorphism.
Genetic structure. The FST statistic for the total population was 0.059 (p=0.001) indicating a low differentiation between the subpopulations. The FIS value obtained for the total population was 0.285 (p=0.010), indicating a slight tendency towards endogamy within the subpopulations, in the total population also seen a slight tendency towards endogamy, represented in the FIT value obtained, which was 0.328 (p=0.10).
The mean flow value of genes represented as the number of migrants (Nm) was 4 individuals per generation; this value represents the movement of individuals or gametes among the subpopulations, promoting a genetic interchange among subpopulations.
Genetic differentiation. The paired FST were between 0.006 and 0.174 (Table 4), presenting significant differences indicating degrees of differentiation between the subpopulations from low to medium. The studied subpopulations can be grouped in two branches; the subpopulations of Bello and Belmira are those that are farthest from the genetic tree, while the subpopulations of Entrerrios, La Unión, Medellín and San Pedro de los Milagros show less genetic differentiation (Figure 3).
Linkage disequilibrium. For the analyzed Holstein population, the most frequent combination of genotypes was the double heterozygote AGAT with a frequency of 24.45%, while the less frequent combination of genotypes was the double homozygote AAAG with a frequency of 1.31% (Table 5).
The D and r2 values per population were not significantly different from zero (data not shown). The total population showed D=0.016 and r2=0.066 but these values were not significant (p>0.05), indicating that the presence of alleles in one locus is independent of the presence of alleles in another locus.
Association of INHA-FSHR genotypes with economically important traits.
Descriptive analysis. The PAJUS mean for the Antioquia Holstein population was 5.588±1.492 L/lac, with PP and PG at 3.06±0.26 and 3.89±0.46 respectively. For SCS a mean of 4.19±1.01 was found. The mean for IEP was 414±85 with 1.67±1.17 SPC (Table 6).
Association of INHA-FSHR genotypes with productive traits.
Milk production. The L, H, FSHR variables with H*AP, H*MP y H*AP*MP interactions had a highly significant effect (p<0.01). The other variables did not have a significant effect on the trait (p>0.05).
Protein percentage. The PREAL had a significant effect on this trait (p<0.05), the other variables did not have a significant effect (p>0.05).
Fat percentage. None of the variables included in the model presented a significant effect on the trait (p>0.05).
Association of INHA-FSHR genotypes with health traits.
Somatic cells scores. None of the variables included in the model presented a significant effect on this trait (p>0.05).
Association of INHA-FSHR genotypes with reproductive traits.
Calving interval. The AP*MP interaction had a significant effect on the trait (p<0.05). The other variables did not have a significant effect (p>0.05).
Services per conception. None of the variables included in the model presented a significant effect on this trait (p>0.05).
The mean of each trait for each combination of genotypes is shown in table 6.
DISCUSSION
The A192G and A-320T polymorphisms have been studied in other populations, such as the Holstein China population (5,7,9). For cows of the Holstein China population, have reported that the A and G alleles of SNP A192G have a frequency of 0.55 y 0.45 respectively, similar to the frequencies found for the Antioquia Holstein population. In the Holstein cows of both populations, the most frequent genotype was AG, however for the population of Holstein China bulls the most common genotype was the AA (5,7). For the Holstein China bulls a frequency of 0.691 was reported for the A allele and 0.309 for the T allele (5,7).
Respecting the A-320T polymorphism, for the Holstein China population a frequency was found for the A and T alleles of 0.301 and 0.699 in females, while in males it was 0.321 and 0.679, respectively. For this population, the most common genotype is TT, in males and females, while in Antioquia is more common the heterozygous (7,9).
In spite of the fact that in the total population a greater number of heterozygote individuals was found than was expected, this difference is not significant, since the total population is found in HWE. Only two populations, La Unión and Entrerrios, were not in HWE, the first due to a significant increase of heterozygote animals, indicating a tendency to exogamy, and the second due to a significant reduction of the same, showing a tendency towards endogamy. Within the populations there are several factors that can generate HWE deviations, such as selective pairing, endogamy, population structure and selection, but endogamy and population structure are the main factors that generate HWE deviations due to small differences in the allelic frequencies between subpopulations (11). However, for populations in La Unión and Entrerrios, it is expected that HWE deviations are due to selection processes on traits that can be associated with these polymorphisms, causing an indirect selection and therefore affecting the allelic and genotypic frequencies.
The endogamy coefficient of a subdivided population (FIT) can be divided using Wright’s F statistics between the component due to non-random pairing within subpopulations (FIS) and the subdivision between populations (FST) (13). For the analyzed population, the FIT value obtained (0.328) reflects a low tendency to endogamy; in the same way, the FIS value obtained (0.285) indicates a slight tendency to homozygosity due to non-random pairing between individuals in each subpopulation. The FST value for the total population (0.059) indicates that a moderate differentiation between the populations exists, which represents a reduction in the heterozygosity due to population subdivision.
The Wright FST statistic measures the subdivision of the population and determines if differences exist in the allelic frequencies among the subpopulations. The populations that showed a greater degree of differentiation were Medellín and Belmira, while the populations in La Unión and San Pedro had less differentiation. This can be explained by the technical level implemented in each of these municipalities, with higher levels in the municipalities of Medellín, San Pedro and La Unión in comparison to Belmira, and therefore among the populations with a higher technical level; in terms of genetic improvement strategies that are implemented, mainly artificial insemination, the FST values are less than 0.05, indicating little or no differentiation. Due to this differentiation, the subpopulations in Antioquia do not act like one population.
The structure and differentiation of the Holstein population in the department of Antioquia has also been evaluated using polymorphisms of different genes such as lactoferrine (LTF) and the bovine growth hormone (bGH). Based on the LTF gene, the Holstein cow population was found to have HWE, and at the subpopulation level only the municipality of San Pedro de los Milagros presented HWE deviations, possibly due to an intense selection, since this is one of the most important municipalities in the dairy sector in the department (17). This investigation reports for the population a FIS=-0.0717, indicating a tendency to exogamy. Upon evaluating this population based on the bGh gene, it was also found to be in HWE. Although the total population reported a FST=0.0068, a certain degree of differentiation was found among the subpopulations, the municipality of Marinilla being the one that presented the greatest genetic differentiation (4).
Nm represents the absolute number of migrant organisms that enter each subpopulation in each generation. This flux of individuals causes a rapid decrease in the fixation index. Thus, migration exercises an important force against genetic divergence between subpopulations (18). For the population analyzed, a total of four migrants per generation was found. Although it is affirmed that if Nm>1 the subpopulations evolve as just one population, it is more correct to affirm that there is no population subdivision if Nm>10 (11). This could explain the genetic differentiation found among subpopulations in the department of Antioquia; although there is genetic flux between them, it has not been sufficient to homogenize the allelic frequencies, although it could be thought that the populations are in a process of homogenization. In other studies (4, 19), the number of migrants that has been reported is much greater than what was found in this investigation. Based on a SNP, the growth hormone gene (bGH) was reported for the Holstein population in Antioquia at Nm=36 (4), while based on the SNP of the lactoferrine gene (LTF), Nm=25 has been reported for the same population (19). This high value could be due to implementing artificial insemination that eliminates geographic barriers and increases the genetic flux between populations (4,19).
The polymorphisms analyzed are found in linkage equilibrium, that is, the observed frequency to combine genotypes does not significantly differ from the expected frequency. In this way, it can be asserted that the presence of each allele for each gene analyzed is independent of the presence of any other. Since in bovines the INHA and FSHR genes are found in different chromosomes, 2 and 11 respectively, these have a maximum recombinant rate, that is, 0.5, confirming their independent segregation according to the second law of Mendel (19).
The mean milk yield was greater than values reported in other investigations (4.482 L/lac ) for the Holstein Antioquia population (20). However, in other studies a greater value (7.155 L/lac) for milk production has been found for this population (21). The PP found in this investigation was less than that found by other investigators (3.16%), and on the other hand, the mean found for PG was greater than the value reported in literature (3.37%) (20). Regarding reproductive parameters, the SPC and IEP averages were very similar to the average reported in a previous investigation, where a mean of 1.67 and 453 days, respectively, was found (22).
The interaction of the studied polymorphisms did not present a significant effect on the majority of the traits that are of Zootechnical interest, however this interaction makes the A-320T polymorphism have a significant effect on the PAJUS, which had not been discovered when analyzing it independently (data not shown). For A-320T polymorphism, the AA individuals evidenced a greater productive level (5.902 L/lac), with 193 and 381 L/lac more than the AT and TT individuals respectively. The AA individuals also showed a lesser IEP value in comparison with the other genotypes (403 days) (data not shown). Since AA individuals present greater reproductive efficiency, reflected in a lower IEP, it would be expected that these individuals present shorter lactation periods, which when they adjusting to 305 days show improvement and a greater production level is obtained. When analyzing the combination of alleles for these genes, the GGAA and AAAA individuals are those that present the greatest productive levels, confirming that the A allele for FSHR is associated with greater production. Regarding the milk solid content, the AGTT individuals present the greatest percentages of fat and protein. In parallel studies that were done, it was found that the G allele in INHA is associated with greater PG, while the FSHR T allele is related to greater PG and PP, since the FSHR T allele is also related to a greater volume of milk, causing a minor dilution effect on milk solids.
A lower IEP value was found in AAAT individuals, while a greater value was found in GGAA, a difference that could be due to INHA alleles, since the A allele is associated with a lesser IEP while GG individuals have greater values for this trait. The lesser SPC number was for GGTT individuals and the greater one for AGAT, but it cannot be stated that one of the analyzed alleles presents a tendency to increase or decrease the value of this trait.
Based on this information, we can conclude that selection based on the phenotype in the population of Holstein Antioquia cows has not been sufficiently strong to generate Hardy-Weinberg equilibrium deviations or diminished in any notable way the frequency of any alleles for the studied polymorphisms, and therefore a great diversity exists among the populations. This genetic diversity greatly important, since individuals can respond adequately to diverse environmental challenges, and additionally it is the starting point to begin genetic improvement programs that seek to increase productive and reproductive efficiency in animal production systems. More studies are needed to determine if these polymorphisms can be useful in molecular marker selection programs and it is recommended that better control over environmental factors be exercised in order to make it easier to detect the effect these polymorphisms could have on the evaluated zootechnical traits.
REFERENCES
1. Boichard D, Maignel L, Verrier E. The value of using probabilities of gene origin to measure genetic variability in a population. Genet Sel Evol 1997; 29:5–23.
2. Muasya T, Peters K, Kahi A. Breeding structure and genetic variability of the Hosltein Friesian dairy cattle population in Kenia. Anim Genet Resour 2013; 52:127–132.
3. Deb R, Chakraborty S, Singh U. Molecular Markers and Their Application in Livestock Genomic Research. J Vet Sci Technol 2012; 3(2):7579.
4. Rincón JC, López A. Echeverri JJ. Estructura y diversidad genética en vacas Holstein de Antioquia usando un polimorFISmo del gen bGH. Rev. MVZ Córdoba 2013; 18(1):3346–3354.
5. Tang KQ, Li SJ, Yang WC, Yu JN, Han L, Li X et al. An MspI polymorphism in the inhibin alpha gene and its associations with superovulation traits in Chinese Holstein cows. Mol Biol Rep 2011; 38(1):17–21.
6. National Center for Biotechnology information. Reference SNP (refSNP) Cluster Report rs41257116. 2013. Disponible en: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=41257116
7. Sang L, Du QZ, Yang WC, Tang KQ, Yu JN, Hua G et al. Polymorphisms in follicle stimulation hormone receptor, inhibin alpha, inhibin beta A, and prolactin genes, and their association with sperm quality in Chinese Holstein bulls. Anim Reprod Sci 2011; 126(3-4):151–156.
8. National Center for Biotechnology information. Reference SNP (refSNP) Cluster Report rs43676359. 2013. Disponible en: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=43676359.
9. Yang WC, Li SJ, Tang KQ, Hua GH, Zhang CY, Yu JN et al. Polymorphisms in the 5’ upstream region of the FSH receptor gene, and their association with superovulation traits in Chinese Holstein cows. Anim Reprod Sci 2010; 119(3-4):172–177.
10. Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16(3):1215.
11. Nielsen R, Slatkin M. An Introduction to Population Genetics. Theory and Applications. Sunderland, Massachussets. USA: Sinauer Associates, Inc. Publishiers, 2013, p. 287.
12. Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012; 28(19):2537–2539.
13. Piñero D, Barahona A, Eguiarte L, Rocha A, Salas R. La variabilidad genética de las especies: aspectos conceptuales y sus aplicaciones y perspectivas en México. En: Díaz P, Morales E, Zizumbo-Villarreal D, editores. Capital Natural de México. México; 2008; p. 415–435.
14. Hall B. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 2013; 30(5):1229-1235
15. Rodríguez-Zas SL, Gianola D, Shook GE. Evaluation of models for somatic cell score lactation patterns in Holsteins. Livest Prod Sci 2000; 67(1-2):19–30.
16. SAS Institute Inc., Cary, NC, USA, 2009.
17. Rodríguez N, López A, Echeverri JJ. Diversidad Genética de un PolimorFISmo del Gen de Lactoferrina Bovino (LTF) en una Población de Vacas Holstein de Colombia y su Asociación con Componentes de la Leche (Resultados Preliminares). Actas Iberoam Conserv Anim 2011; 1(1):151–153.
18. Hartl DL, Clark AG. Principles of population genetics. 4th ed. Sunderland, Massachusetts: Sinauer Associates, Inc. Publishiers, 2007.
19. Rodríguez N, López A, Echeverri JJ. Estructura genética poblacional del gen lactoferrina bovino en vacas Holstein del departamento de Antioquia. Rev MVZ Córdoba 2013; 18(1):3355–3361.
20. Echeverri JJ, Salazar V, Parra JE. Análisis comparativo de los grupos genéticos Holstein, Jersey y algunos de sus cruces en un hato lechero del Norte de Antioquia en Colombia. Zootec Trop 2011; 29(1):49–59.
21. Corrales J, Cerón-Muñoz M, Cañas J, Herrera C, Calvo S. Parámetros genéticos de características de tipo y producción en ganado Holstein del departamento de Antioquia. Rev MVZ Córdoba 2011; 17(1):2870–2877.
22. Madrid S, Echeverri JJ. Association Between Conformation Traits and Reproductive Traits in Holstein Cows in the Department of Antioquia – Colombia. Rev Fac Nac Agron Medellín 2014; 67(2):7311–7319.