Detection and molecular characterization of Cryptosporidium spp. in dairy calves from the Valdivia province, Chile
Detección y caracterización molecular de Cryptosporidium spp. en terneros de lechería de la provincia de Valdivia, Chile

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Show authors biography
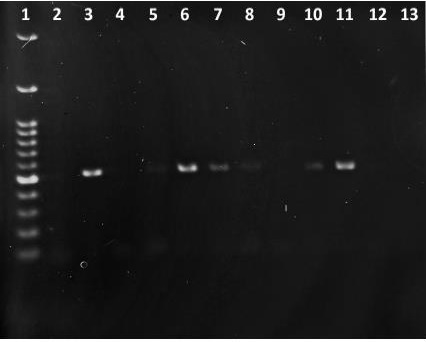
Objective: To detect acid-fast oocysts of Cryptosporidium spp. using the Ziehl-Neelsen stain (ZN) and apply molecular tools with the 18S rARN and 60 kDa glycoprotein (gp60) genes to characterize the species and subtypes present in the Valdivia province. Material and methods: 275 samples of calves from 15 dairy farms located in the Valdivia province were analyzed by ZN stain. Samples that were positive to ZN were then submitted to PCR processing aiming as target the 18S rARN and gp60 genes, with subsequent sequencing of the latter. Results: Thirty samples were positive to ZN, of which 46.6% (14/30) were confirmed by PCR amplification for 18S rARN and from those, 42.8% (6/14) also amplified the gp60 target using PCR. Subsequent sequencing of this gene showed the presence of C. parvum in 100% of the isolates and the subtypes IIaA15G2R1 (3/6), IIaA14G1R1 (2/6) and IIaA17G1R1 (1/6). Conclusions: Subtype IIaA15G2R1 was predominant, being found in two boroughs of the province. The subtype IIaA14G1R1 was found in two farms located in the same borough and, to our knowledge, this is the first time that this subtype has ever been reported in cattle in both Chile and the continent. Finally, subtype IIaA17G2R1 has been found sporadically in calves just like in the present study.
Article visits 857 | PDF visits























